 |
|
Table of Contents:
Details of the VOLCANO'09 Challenge:The target of the challenge is three-dimensional change analysis of pulmonary nodules in CT images. The focus of the challenge is not directly on segmentation itself (which tells us little of the underlying disease) but rather the change in size of the lesion recorded on two time-separated images. This size change is a critical measurement for (a) diagnosing cancer and (b) evaluating response to therapy. One of the most important indicators of malignancy is the relative change in size of a nodule over a period of time. The critical issue for the challenge, the precision of size change measurement, is needed to establish the minimum time delay between sequential scans and the associated magnitude of the measurement required to determine malignancy or response to therapy. Most evaluation methods for CAD systems, including challenges, involve a ground truth established be experts. However, for the task of lesion size estimation it is well known that there is a large amount of variation or disagreement in expert size estimations. Further, it has not been established that experts manual estimations are superior to automated measurements. In this challenge, while the change in size of lesions will be reviewed by experts, we will explore the issue of ground truth through the submitted responses to the challenge. Motivation for the studyCurrent approaches to quantification of nodule volume change measurement exhibit two main problems that complicate their direct comparison. First, these methods require large unified database of both stable and growing nodules. Second, there is no single commonly used evaluation technique that would assess the measurement quality of a particular method. Therefore we invite interested parties to take part in this unique study that address both of these issues by providing a single evaluation image dataset and a common methodology for assessing the quality of the measurement algorithm. Goals of the studyBy conducting this challenge we are trying to achieve the following goals that we believe will be beneficial for the lung CAD research community:
RulesOrganization of this study and maintaining this website is a large effort. We ask everyone who decided to participate in the challenge to read and accept the following rules.
Data DescriptionThe image data used in the study was acquired for the Public Lung Database to to address drug response and was provided by the Weill Cornell Medical College. Cases were selected that contained at least one nodule of solid consistency which was present in at least two scans with a whole-lung field of view including the entire nodule. Only nodules visible on at least three slices on both scans were included. 53 total nodules are available to the challenge in this way. Evaluation DatasetThe evaluation dataset consists of 49 nodules divided into three categories. The first category consists of 27 nodules visible on two scans of 1.25 mm slice thickness, have little observed size change, and a range in diameter from approximately 4 - 24 mm. These cases span the sizes of most interest for nodule growth measurement and represent good quality scans. The second category of nodules included 13 nodules imaged on either two 2.5 mm scans or one 1.25 mm scan and one 2.5 or 5.0 mm scan to examine the effect of slice thickness on the performance. The nodules ranged in size from approximately 8 – 30 mm. The third category consists of an additional 9 nodules on two 1.25 mm scans, but a large size change; these nodules ranged in size from approximately 5 – 14 mm. The approximate size distribution of nodules in the evaluation dataset is shown in the plot below: 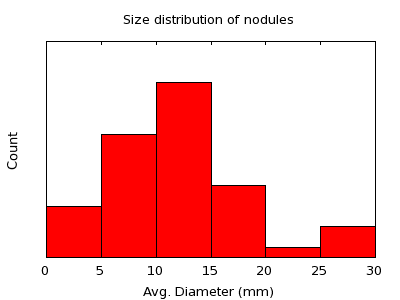 The sizes used to produce this histogram are only estimates. Example DatasetFour nodules are provided as examples spanning the three categories described above. These nodules will not be considered in performance evaluation. Data PreparationAll of the images for this challenge are made available in DICOM format with all patient information removed. The original dates have been removed from the scans and replaced with dates corresponding to a time interval of 100 days between each pair of scans, with the order of the scans randomized. Scans were clipped in the axial direction, and where possible, the five slices above and below the region containing the nodule were included in the clipped scan. Nodule LocationsFor each pair of nodules, the following information to locate the nodules is provided in a CSV file:
The nodule locations are in the approximate center of the nodule, on the slice with the largest area. If your algorithm requires a seed point, these are the points that should be used. If you need to use a different seed point, please indicate these seed points in your submission. There are two files, one for the example dataset and one for the evaluation dataset. Format of the submissionThe critical information that must be included with each result submission is the proportional change in size of the lesion between the two scans relative to the size of the lesion in the first scan. If the measurement system measures the volume of the lesion in the first scan as V1 and the volume of the lesion in the second scan as V2 then the required number is (V2 - V1)/V1. It is recognized that some systems do not need to explicitly evaluate volumes in order to estimate change in size.
Each team must provide a spreadsheet in either CSV or Excel format for
only those cases in the evaluation dataset with at least the following columns,
where V1 and V2 are the volumes (mm^3) of the nodule on the first and
second scans respectively: It is quite possible that some methods may not work for some of the cases. For this situation please provide the case ID but leave the other values in that row empty. Requirements for the supporting PDF documentationAny submission of the results should be accompanied by a PDF document describing the change measurement methodology. This shoudl include a description of any parameter settings used to create the results and any user interaction should be clearly explained. Alternatively, a copy of a published paper may be submitted. Submissions without a description of the method will be rejected. There is no specific style requirement, however the following items would typically be mentioned in the document:
Workshop information
Details of the workshop pertinent to this challenge will appear
here as they become available. The format of the paper to be
submitted will be posted when available at the workshop
website.
|
||||||||||||